Data Documentation
Data documentation will ensure that your data will be understood and interpreted by any user. It will explain how your data were created, the context for the data, the structure of the data and their contents, and any manipulations to the data.
What is important to document?
- Context of data collection
- Data collection methodology
- Structure and organization of data files
- Data validation and quality assurance
- Data manipulations through data analysis and use conditions
Data-level documentation
- Variable names and descriptions
- Definition of codes and classification schemes
- Codes of, and reasons for, missing values
- Definitions of specialty terminology and acronyms
- Algorithms used to transform data
- File format and software used
Example Readme Files
README.txt files are text files that allow researchers to keep textual notes on their digital data files. These README.txt files contain documentation that is easily and immediately understandable. They allow you to add notes about the organization and content of your digital files and folders, which helps other researchers or colleagues to navigate the data. Ideally, README.txt files are kept at the top level of a project folder to provide the purpose of the project, the relevant summary and contact details, and general organization of files. Think of them like the first page of your lab notebook.
- All-purpose, structured README.txt: example 1 and example 2 use an Open Science Framework template
- Comprehensive README.txt: Part of a survey dataset, this readme documents the data analysis process and explains each file and folder. The dataset includes license files. The dataset has also been published in Zenodo, capturing many of the metadata fields suggested in the Metadata section below.
- Simple README.txt: For easy data management, add a simple README.txt to your project folders
Metadata
Metadata describes the origin, purpose, time, geographic location, creator, access, and terms of use of the data. Information in the metadata is used to retrieve and index data in a repository or archive, and enables citation of the data. Metadata can be harvested for data sharing through the Open Archives Initiative Protocol for Metadata Harvesting (OAI-PMH).
There are a variety of metadata standards, usually for a particular file format or discipline. Some examples include the following:
- Human Pathogen and Vector Sequencing Metadata Standards
- Metadata Standards Catalog for Clinical Medicine
- Data Curation Centre’s List of Metadata Standards
- Clinical Data Interchange Standards Consortium
- NIH Common Data Elements
An excellent guide to medical metadata for research data is Johns Hopkins Guide to Documenting Research Data.
Consult these directories for comprehensive lists and tools of discipline-specific metadata.
The Wood Library can help you select the most appropriate metadata standard to use. Contact Wood Library.
When creating metadata, a best practice is to use controlled vocabulary or standard terminology for your discipline. Using a controlled vocabulary or an authority list will help in the retrieving and indexing of your data.
Consider keeping metadata records in a spreadsheet, CSV file, or tab-delimited file. Additional information to interpret the metadata, such as explanations of variables, codes, acronyms, abbreviations, or algorithms, should be included as accompanying documentation.
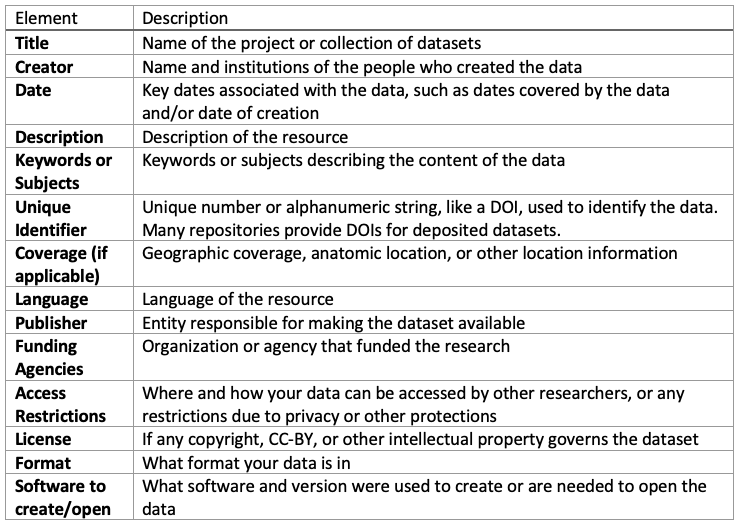
Suggested Metadata Elements
The Wood Library suggests the following metadata elements. In their simplest form, these can be included as part of a README.txt file. The Open Science Framework README.txt template contains a minimal set of elements.